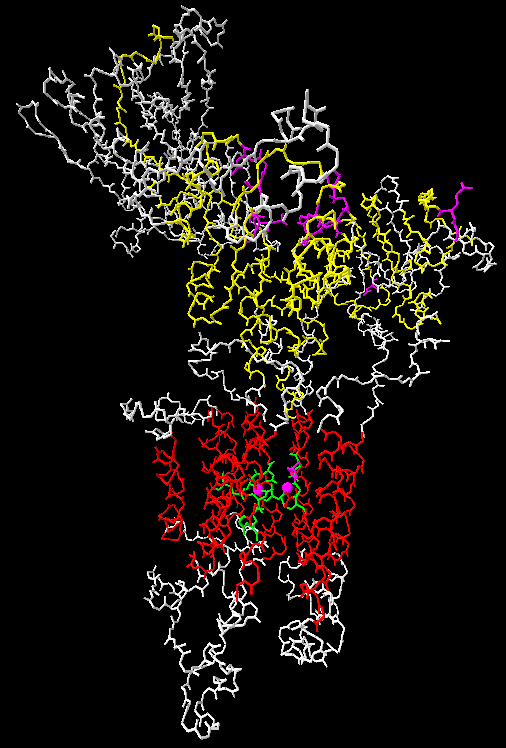
 The structure of the P2A ATPase SERCA1 from rabbit has been solved three
times, in two different conformations and with a bound ATP analog. The structures were
published in Nature:
The structure of the P2A ATPase SERCA1 from rabbit has been solved three
times, in two different conformations and with a bound ATP analog. The structures were
published in Nature:
Toyoshima, C., Nakasako, M., Nomura, H., and Ogawa, H. (2000) Crystal structure of the
calcium pump of sarcoplasmic reticulum at 2.6 Å resolution.
Nature 405: 647-655,
Toyoshima,
C., and Nomura, H. (2002) Structural changes in the calcium pump accompanying the
dissociation of calcium.
Nature 418: 605-611, and
Toyoshima,
C., and Mizutani, T. (2004) Crystal structure of the calcium pump with a bound ATP analogue.
Nature 430: 529-535.
The structures can also be found at the PDB databank with the accession numbers 1SU4, 1IWO and 1VFP.
The structure seen here is of 1SU4 and was made with Swiss-PDBviewer which can be downloaded from the ExPASy site.
TM regions are red, conserved segments are yellow, universally conserved residues are violet, and calcium coordinating residues are green. The two calcium ions are seen as violet spheres.
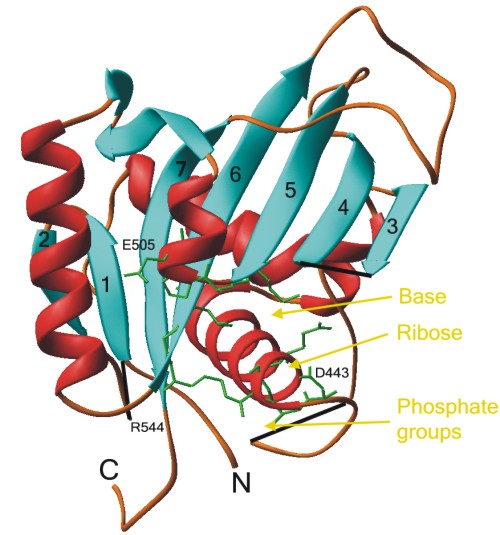 The structure of the N-domain of the pig P2C ATPase
The structure of the N-domain of the pig P2C ATPase  2 subunit has also
been solved, and is shown here in a ribbon diagram kindly provided by K.O. Håkansson.
2 subunit has also
been solved, and is shown here in a ribbon diagram kindly provided by K.O. Håkansson.
Suggested binding sites for the adenine base, ribose group and the phosphate groups
of ATP are shown in yellow. Amino acid residues from the conserved motifs GDASE446 (right),
KGAPE505 (top), and RXL546 (left) are shown as green stick-and-balls.
The random coil structure and the C-terminus is the His6 tag.
Gaps in the structure have been mended with black lines for clarity.
The structure was published in: Håkansson, K.O. (2003) The crystallographic structure of Na,K-ATPase N-domain at 2.6Å resolution. J. Mol. Biol. 332: 1175-1182,
and has the PDB accession number: 1Q3I.
I hope once to find time to incorporate as much as possible of the structural information in the alignments of the different subfamilies of P-type ATPases.
Go to Front page
 The structure of the P2A ATPase SERCA1 from rabbit has been solved three
times, in two different conformations and with a bound ATP analog. The structures were
published in Nature:
The structure of the P2A ATPase SERCA1 from rabbit has been solved three
times, in two different conformations and with a bound ATP analog. The structures were
published in Nature:  The structure of the N-domain of the pig P2C ATPase
The structure of the N-domain of the pig P2C ATPase  2 subunit has also
been solved, and is shown here in a ribbon diagram kindly provided by K.O. Håkansson.
2 subunit has also
been solved, and is shown here in a ribbon diagram kindly provided by K.O. Håkansson.