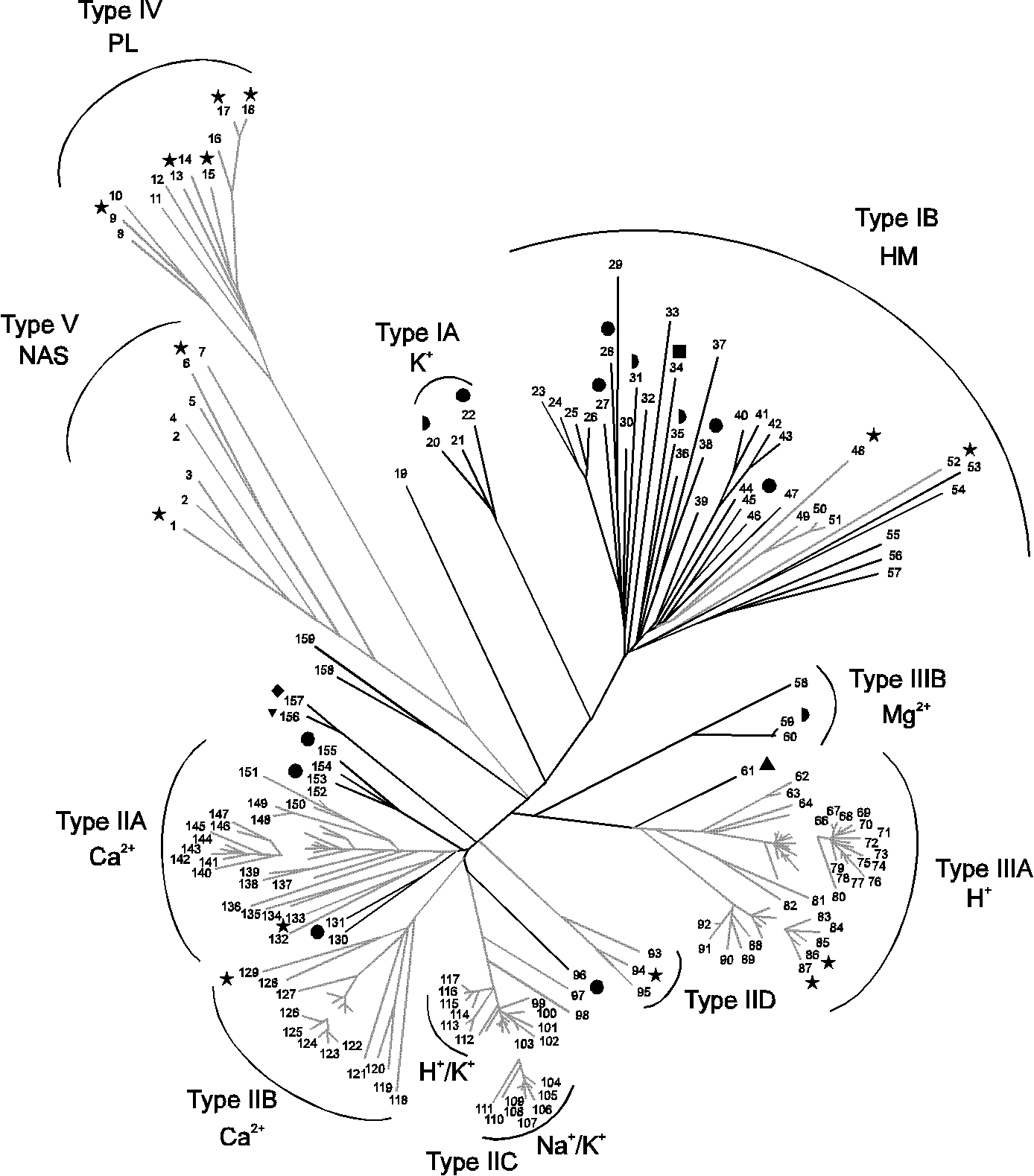
Phylogenetic tree of P-type ATPases
The phylogenetic is based on core sequences from 159 P-type ATPase sequences present in the EMBL and SWISS-PROT databases July 1, 1997. Further 53 ATPase sequences present in the databases were not included in the tree, as they showed more than 90% sequence identity (full-length proteins) to one of the included sequences. The tree was constructed with the Phylip program package using the Protdist and Fitch programmes. Some areas (which are not connected to the rest of the tree) have been enlarged 40 % to clarify the distribution of species. When the substrate specificity of the ATPases present in each family is known, it corresponds in all cases to the name of the family. The numbers of the sequences correspond to the numbers defined in
Axelsen, K.B. & Palmgren, M.G. (1998) J. Mol. Evol. 46: 84-101, Table 1. The numbers are always shown together with the corresponding sequences in this database. The black branches show ATPases originating from Bacteria and Archaea, and the grey branches show ATPases originating from Eukarya. The P-type ATPases from the fully sequenced organisms are shown with the following symbols:  : Escherichia coli;
: Escherichia coli;  : Haemophilus influenzae;
: Haemophilus influenzae;  : Methanococcus jannaschii;
: Methanococcus jannaschii;  : Mycoplasma genitalium;
: Mycoplasma genitalium;  : Mycoplasma pneumoniae;
: Mycoplasma pneumoniae;  : Saccharomyces cerevisiae;
: Saccharomyces cerevisiae;  : Synechocystis PCC6803. The abbreviations are HM: heavy metals; NAS: no assigned specificity; PL: phospholipids.
: Synechocystis PCC6803. The abbreviations are HM: heavy metals; NAS: no assigned specificity; PL: phospholipids.
To view a larger version of the tree click here.
Go to Front page

